❄️ How do plants adapt their metabolism to survive cold stress?
🌿 Cold stress severely impacts plant growth & secondary metabolism, posing a challenge for crops like Dendrobium catenatum.
🧬 Here researchers identified 62 DcbZIP genes in D. catenatum, a large family of transcription factors (TFs) involved in cold stress responses.
🌡️ Among these, 58 DcbZIPs were highly responsive to cold stress, especially DcbZIP3, DcbZIP6, & DcbZIP28 in leaves & stems.
🔍 Gene analysis showed that these DcbZIPs contain the conserved bZIP domain & are localized in the cell nucleus, driving gene regulation.
🧩 DcbZIP6 negatively correlated with PAL2, a key player in flavonoid metabolism, while DcbZIP28 showed negative links to polysaccharide metabolism genes like PFKA1, ALDO2, & SCRK5.
🌱 This suggests that DcbZIP6 mainly regulates flavonoid metabolism, while DcbZIP28 controls polysaccharide metabolism under cold stress.
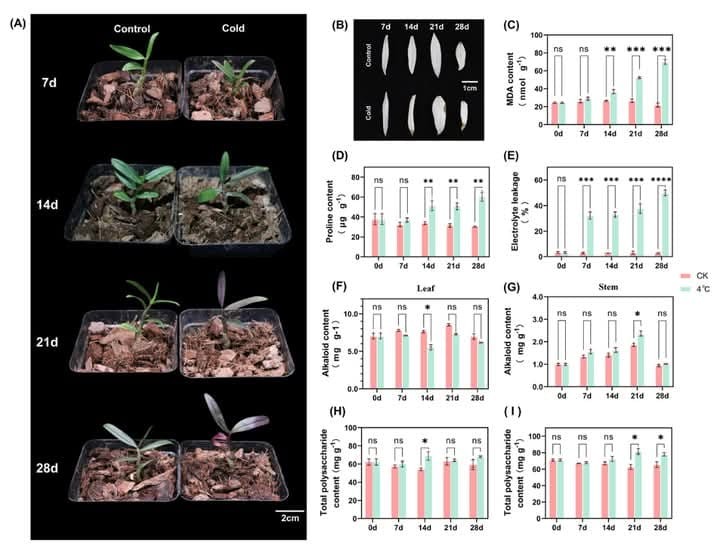
❄️ Interestingly, cold stress also increased total polysaccharide & alkaloid contents in D. catenatum stems, highlighting metabolic shifts.
🔬 These insights reveal how the DcbZIP gene family helps plants adjust metabolism to withstand cold, offering new targets for stress-resilient crops.
💡 Could manipulating these transcription factors unlock new ways to protect plants from harsh climates?